Scientists Develop Novel Raman Method to Monitor Beer Fermentation at Single-Cell Level
Breweries typically monitor fermentation by measuring the broth: alcohols, esters, acids and residual sugars are quantified with chromatography-based assays that are reliable but time-consuming, and they report only batch averages. A new study shows that a rapid, label-free readout from individual yeast cells can be converted into many of those same process metrics—while also revealing cell-to-cell variation that bulk tests hide.
In a collaboration between Tsingtao Brewery and the Single-Cell Center at the Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT) of Chinese Academy of Sciences, researchers developed a workflow they call “process ramanomics,” based on spontaneous single-cell Raman spectroscopy.
The study was published on December 9 in Bioresource Technology.
The team tracked an industrial beer fermentation using the lager yeast Saccharomyces pastorianus, sampling a single batch across eight days. At each stage, they collected high-throughput Raman spectra from individual cells (a “ramanome”) and matched those fingerprints to conventional measurements of 43 extracellular phenotypes in the fermentation medium.
Using multivariate regression, the researchers showed that ramanomes could predict 19 extracellular phenotypes with strong performance. These included four higher alcohols, four esters, four amino acids, two organic acids, and four mono- and disaccharide substrates, plus the alcohol-to-ester ratio, a commonly used indicator linked to flavor balance. In effect, one fast cellular measurement can stand in for multiple chemical assays, without losing single-cell detail.
Because the models output cell-level predictions, the researchers also tracked phenotypic heterogeneity over time. Different metabolite classes displayed distinct heterogeneity trajectories, and for several phenotypes higher heterogeneity tended to accompany lower metabolite levels—suggesting that dispersion among cells may be a useful process-state indicator.
To support mechanistic hypotheses, the authors introduced Intra-Ramanome Correlation Analysis (IRCA), which extracts correlation networks linking intracellular Raman features, extracellular metabolites and substrates. Their analysis highlighted carbohydrates as the most dynamic intracellular pool, and pointed to protein-associated signals as closely connected to alcohol and ester formation early in fermentation.
“Instead of waiting for tank chemistry to drift before we notice it, we can read the cells directly and infer multiple process outputs from their metabolic fingerprints,” said Prof. XU Jian from the Single-Cell Center, a co-corresponding author.
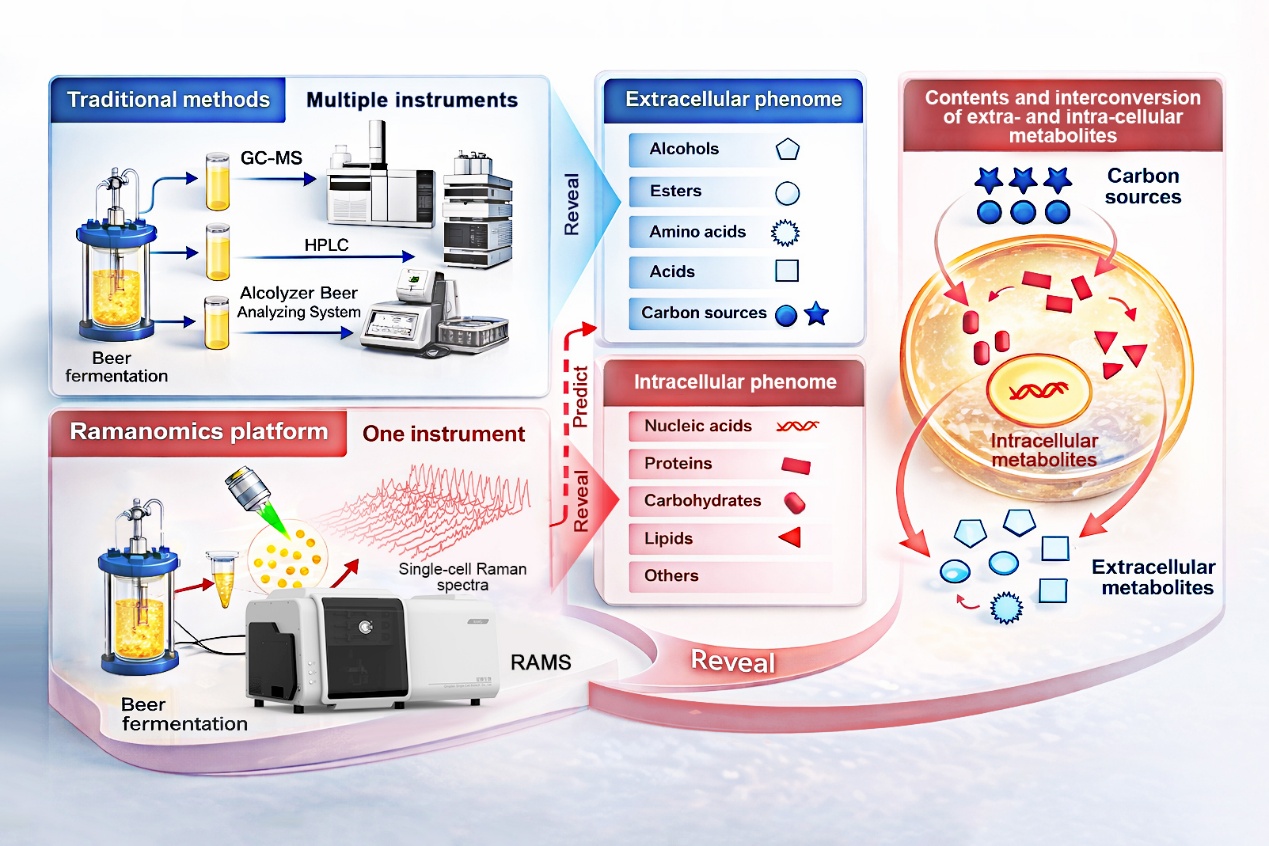
Overview of “process ramanomics”. Single-cell Raman fingerprints collected across fermentation provide a fast, label-free window into brewing progress with single-cell resolution.
(Text/Image by LIU Yang)