Raman Flow Cytometry Efficiently Screens Lipid-Rich Yeast Mutant for Functional Fatty Acid Production
Researchers at the Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT), Chinese Academy of Sciences, have identified a lipid-rich mutant of Saccharomyces cerevisiae using a high-throughput, label-free Raman flow cytometry known as FlowRACS.
The findings, published in Biotechnology for Biofuels and Bioproducts on July 17, open new avenues for microbial production of palmitoleic acid—an omega-7 fatty acid with demonstrated anti-inflammatory and metabolic benefits.
Palmitoleic acid is scarce in conventional oil crops and typically sourced from plants like sea buckthorn and macadamia, which are limited by geography and yield. Although S. cerevisiae naturally accumulates palmitoleic acid, its low total lipid content has hindered its industrial application.
To address this limitation, the researchers applied a combined mutagenesis strategy using zeocin and Atmospheric and Room Temperature Plasma (ARTP) to generate a diverse mutant library. They then used FlowRACS to directly select living yeast cells with elevated lipid signals based on their intrinsic single-cell Raman spectra, without the need for stains or reporters.
This approach led to the identification of mutant strain MU2R48, which achieved a lipid content of 40.26%, representing a 30.85% increase over the parental strain SC018, while maintaining comparable biomass levels.
“FlowRACS system allows label-free, non-invasive selection of lipid-producing cells at single-cell resolution,” said JI Xiaotong, co-first author of the study. “It provides a direct link between phenotype and function without genetic modification or chemical labeling.”
Multi-omics analysis revealed that the enhanced lipid accumulation in MU2R48 was driven by coordinated metabolic rewiring: upregulation of glycolysis, ethanol degradation, and the pentose phosphate pathway increased fluxes of acetyl-CoA and NADPH, while fatty acid degradation pathways were suppressed.
“This metabolic rewiring demonstrates how targeted mutagenesis and advanced screening can synergize to optimize microbial production,” said co-corresponding author Prof. ZHOU Wenjun. “The mutant strain provides a promising chassis for palmitoleic acid biosynthesis.”
Prof. MA Bo, another co-corresponding author, emphasized, “FlowRACS represents a powerful tool for functional strain accumulation and sorting—its label-free, high-throughput design is well-suited for modern biomanufacturing pipelines.”
The study underscores the potential of integrating advanced cell sorting with multi-omics analysis to create high-performance microbial strains without relying on transgenic methods. With its scalable and label-free design, FlowRACS has become a key tool in the microbial biotechnology toolbox.
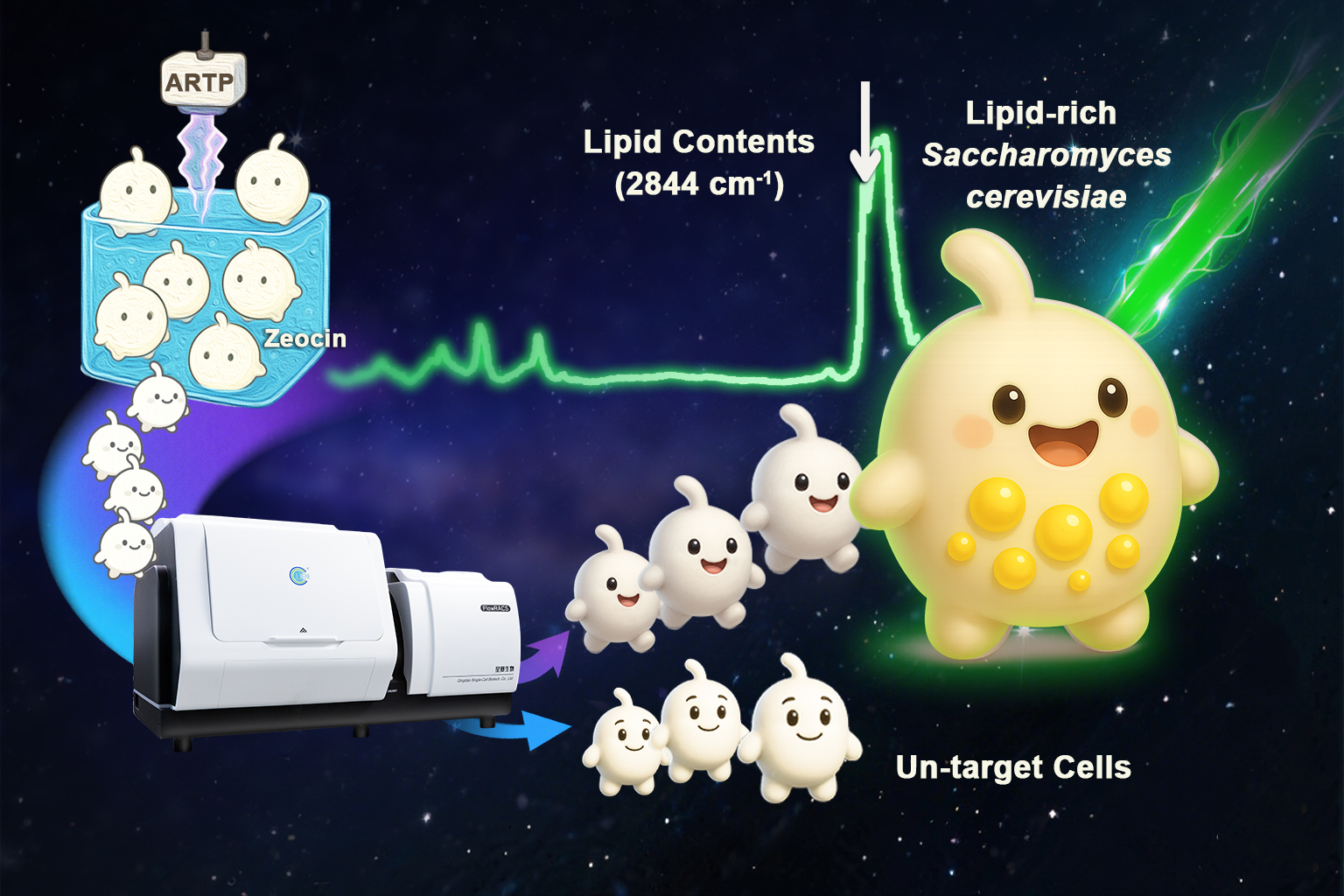
Raman flow cytometry efficiently identifies lipid-rich Saccharomyces cerevisiae mutants from a Zeocin–ARTP-induced library.(Image by QIBEBT)