New "Species-Metabolism" Dual-targeting Technology Unlocks Microbial and Enzyme Resources
A cutting-edge technology developed by Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT), Chinese Academy of Sciences, in collaboration with several renowned institutions, is poised to transform the discovery of microbial species and enzymes with specific in situ metabolic functions.
The team’s findings were published in The Innovation on Jan 17.
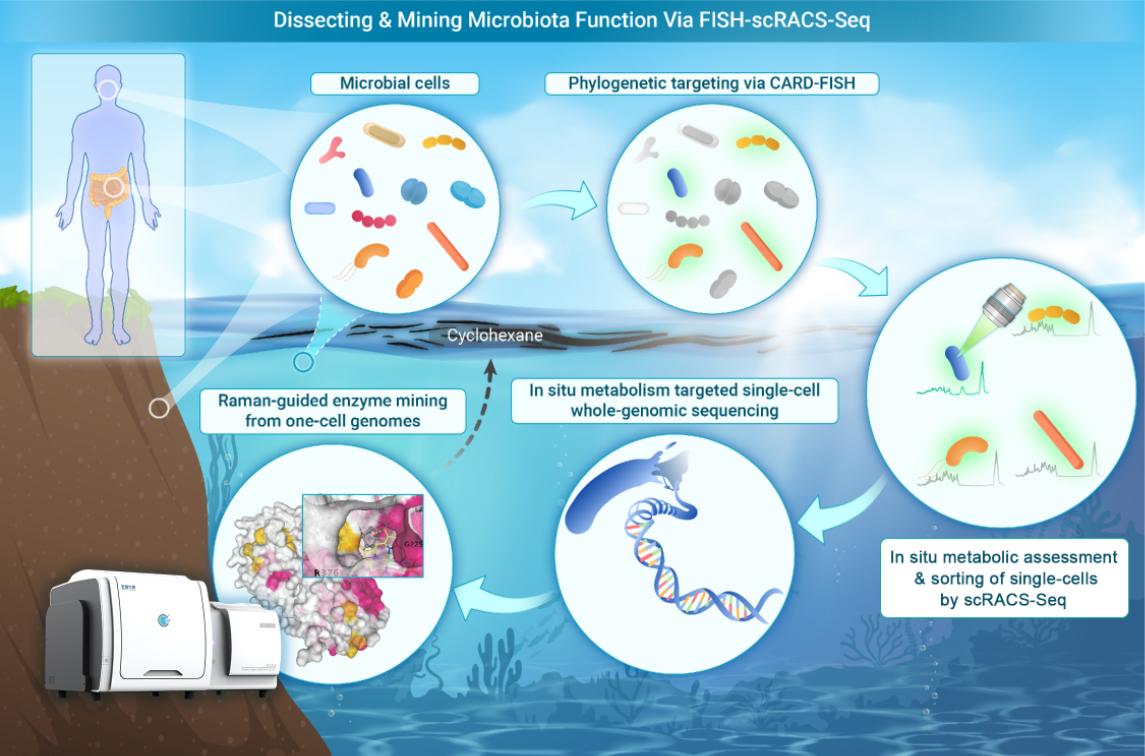
Phylogeny-metabolism dual-directed cell analysis, sorting and sequencing
The new technology, FISH-scRACS-seq (Fluorescence-In-Situ-Hybridization-guided Single-Cell Raman-activated Sorting and Sequencing), leverages the Raman-activated Optical Tweezers-based Cell Sorter developed by Single-Cell Biotech Inc. to combine species-targeting fluorescence in situ hybridization (FISH) with Raman spectroscopy, enabling the direct identification and isolation of functional single cells and their encoding enzymes from environmental samples.
This dual-targeting approach enables researchers to match microbial species to their specific metabolic functions and uncover the underlying genomic resources that drive these processes. The method allows for the high-resolution study of ecological processes, linking species, metabolic abilities, genome sequences, metabolic pathways, and enzyme activities.
“FISH-scRACS-seq is an exciting technology that opens up unprecedented avenues for studying microbial metabolic functions in natural environments,” said Asso. Prof. JING Xiaoyan, first author of the study from Single-Cell Center at QIBEBT. “By overcoming the respective limitations of existing single-cell omics approaches that are either gene-driven (missing function) or metabolism-driven (unable to target specific organisms), FISH-scRACS-seq enables high-resolution analysis of microbial functions, metabolic pathways, and genome sequences directly within natural ecosystems.”
Using this technique, the research team successfully identified cells, pathways and enzymes from γ-proteobacteria that are actively degrading cycloalkanes in situ in marine environments. The analysis revealed a previously unknown P450 enzyme system responsible for cycloalkane degradation, which is crucial for bioremediation in aquatic ecosystems contaminated by hydrocarbons.
Validated through experiments involving complex soil and mixed-culture systems, FISH-scRACS-seq can achieve over 99% single-cell genome coverage, allowing for comprehensive exploration of enzymatic genes and regulatory elements that support in situ metabolic functions of their host cells.
A highlight of the study was the identification of Pseudoalteromonas fuliginea, a bacterium capable of efficiently degrading cyclohexane through a newly identified cytochrome P450 enzyme system (P450PsFu).
This enzyme system converts toxic cyclohexane into non-toxic cyclohexanol. This marks the first step in the degradation pathway and offers insights into marine carbon cycle dynamics. Although P450PsFu is rare in marine microbiomes, its widespread presence - particularly in cold marine ecosystems - opens new avenues for understanding hydrocarbon biodegradation in the oceans.
“When combined with Metagenomic-Wide Association Studies (MWAS), this new tool offers a comprehensive solution for linking ecological characteristics to cellular and enzymatic catalytic functions, making it a powerful resource for biotechnological and environmental application,” added Prof. XU Jian, head of the Single-Cell Center who led the study together with Prof. CUI Zhisong from First Institute of Oceanography, Ministry of Natural Resources of China and Prof. MA Li from Shandong University.
(Text/Image by LIU Yang)