Observation of Protein Conformational Dynamics in Live cells
Protein dynamics through motions of loops, linkers, and hinges can generate distinctive conformations that are important for protein function.
For example, protein folding, recognition, conformation, and enzyme catalysis are all related to the conformational dynamics of proteins.
Most proteins perform their functions in cells. However, there is little research on the conformational dynamics of intracellular proteins, and whether the complex cellular environment affects the conformational dynamics of proteins is an important and unresolved question.
Recently, a research team from the Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT) of the Chinese Academy of Sciences (CAS) has investigated the protein loop conformational dynamics in Escherichia coli ( E.coli ) cells by Nuclear Magnetic Resonance (NMR) Spectroscopy.
The study was published in Science Advanceson July. 21.
In the work, the weak interactions between the protein and surrounding macromolecules in E.coli cells hinder the protein rotational diffusion, which extends the dynamic detection timescale up to microseconds by the NMR spin relaxation method. The loop picosecond to microsecond dynamics is confirmed by nanoparticle-assisted spin relaxation and residual dipolar coupling methods (Figure 1B).
Further investigation characterized the protein loop 1 region with strong flexibility through the sequence parameter S2 (0 represents the most flexible; 1 represents the most rigid), and the linkers between α1 and β3 also have significant flexibility (Figure 1B). The conformational dynamics of intracellular proteins are consistent with the results obtained in aqueous solutions, but there are significant differences in numerical values. The loop interactions with the intracellular environment are perturbed through point mutation of the loop sequence. For the sequence of the protein that interacts stronger with surrounding macromolecules, the loop becomes more rigid in cells. In contrast, the mutational effect on the loop dynamics in vitro is small.
In addition, this study realized the direct measurement of the conformation dynamics of protein loop in cells, and provided direct evidence for the cell environment to change the conformation dynamics of protein loop through weak interaction, which may affect the function of proteins. Furthermore, it highlights the importance of directly studying protein properties and functions within living cells.
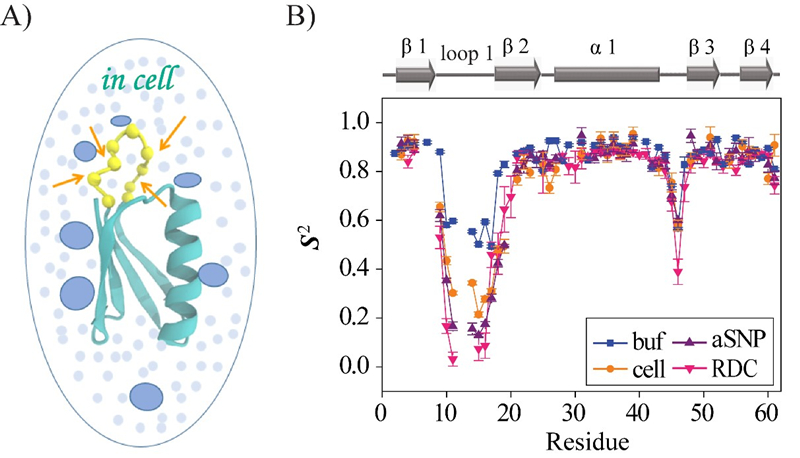
Comparison of conformational dynamics of protein GB3L measured in aqueous solution and in cells(Image by SONG Xiangfei, WANG Mengting and YAO Lishan)
(Text by SONG Xiangfei, WANG Mengting and YAO Lishan)
Contact:
KONG Fengru
Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences
Tel: 86-532-58261072
E-mail: kongfr@qibebt.ac.cn