Tuberculosis is a serious infectious disease. It has caused a huge number of human deaths in history and remains a threat to public health.
The most effective drug to treat tuberculosis is the specific broad spectrum antibiotic. Rifamycins, the first and the most commercially successful antibiotics, show the wide spectrum of antimicrobial activities against tuberculosis bacteria. Their semisynthetic derivatives have long been used as first-line therapies for the treatment of tuberculosis, leprosy, and AIDS-related mycobacterial infections, having saved millions of lives.
Therefore, the mechanisms related to rifamycins biosynthesis has been a research hotspot since their first discovery in 1957. Although a majority of rifamycin B biosynthetic steps have been elucidated, the late biosynthetic steps leading from rifamycin SV to the end product rifamycin B remains elusive to date.
Recently, a collaborative team from Chinese Academy of Sciences (CAS) that consists of Professor LI Shengying Li’s group at Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT), Professors Guoping Zhao’s and Prof. Youli Xiao’s groups at Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences, and Prof. Gong-Li Tang’s group at Shanghai Institute of Organic Chemistry, completely elucidated the long mystery on late rifamycin biosynthesis.
The research, published in Nature Communications, explained the biosynthetic network for the inter-conversion of rifamycin SV, S, L, O, and B, as well as the functionalities and mechanisms of the involved transketolase Rif15 and cytochrome P450 monooxygenase Rif16, thus finally clearing the cloud that has long obscured the late reactions of rifamycin biosynthesis.
In the present study, the late steps of rifamycin biosynthesis were elucidated for the first time by reconstitution of the in vitro activities of transketolase Rif15 and P450 enzyme Rif16, 13C chasing experiments, and resolution of the Rif16 crystal structure. This work revises the previous biosynthetic model that was proposed by Ghisalba et al. 36 years ago (J. Antibiot. 1982, 35, 74-80). Specifically, rifamycin SV can be oxidized to rifamycin S spontaneously. The transketolase Rif15 is responsible for transferring a C2 keto-containing fragment from a 2-ketose to rifamycin S giving rise to rifamycin L with a C-O ester bond. Subsequently, the P450 enzyme Rif16 catalyzes the transformation from R-L to R-O. Finally, R-O is non-enzymatically reduced to R-B by NADPH. All of the mechanisms have been clarified in detail. The elucidation of the network comprising the late steps of rifamycin biosynthesis revealed a unique C-O bond formation reaction mediated by a transketolase and an atypical P450 reaction of ester-to-ether transformation. These uncommon biosynthetic mechanisms adds two new reactions to the toolkit of available biosynthetic tools. It will likely have significant implications for strain engineering relating to the industrial production of rifamycins and the new derivatives thereof.
This work was supported by the Shandong Provincial Key Laboratory of Synthetic Biology, Shandong Natural Science Foundation (ZR2017ZB0207 to W. Zhang and S. Li), the National Natural Science Foundation of China (grants 81741155, 31422002 and 21472204 to S. Li, 31600036 to F. Qi, 21572243 to Y. Xiao, 31430004 and 31670058 to G. Zhao), Chinese Academy of Sciences (grants QYZDB-SSW-SMC042 to S. Li and XDPB0402 to Y. Xiao), and the Science and Technology Commission of Shanghai Municipality (grant 15JC1400402 to Y. Xiao).
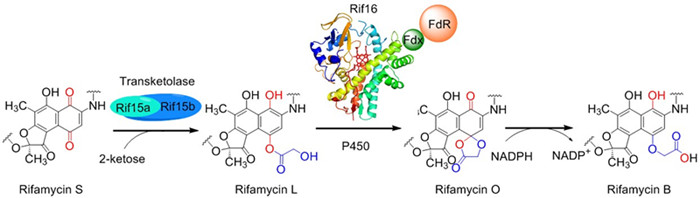
Figure 1. The biosynthetic network of late rifamycin derivatives (Imaged by Qi Feifei)
(Text by Qi feifei, LI Shengying and XIAO Youli)
Contact:
CHENG Jing
Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences
Tel: 86-532-80662647
E-mail: chengjing@qibebt.ac.cn