Genome Editing Platform for Industrial Oleaginous Microalgae
Microalgae are organisms that can capture and assimilate atmospheric CO2 by using sunlight. They then store the solar energy and CO2 in the form of energy-dense molecules such as triacylglycerol (TAG), which can be readily converted to oil. Therefore, interest in microalgae as a scalable solution for clean fuel production and CO2 sequestration has been growing. However, the paucity of enetic and genome engineering tools has greatly hindered mechanistic studies and molecular breeding of energy microalgae. For example, Nannochloropsis spp. are a group of industrial oleaginous microalgae that can be cultured in large scale with sea water. They are of industrial interest due to their ability to grow rapidly under a wide-range of scales, synthesize large amounts of TAGs and high-value polyunsaturated fatty acids (PUFAs) (e.g., eicosapentaenoic acid (EPA)) and tolerate broad environmental and culture conditions. But the lack of reverse genetic engineering tools has hindered efforts to genetically improve key traits such as the efficiency and capacity of carbon fixation and oil production.
In a new study published in the Plant Journal, PhD student WANG Qintao, Dr. LU Yandu and their teammates from the Single Cell Center at Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT) of Chinese Academy of Sciences (CAS) established a novel genome editing platform for the industrial oleaginous microalgae Nannochloropsis oceanica. The platform employs a well-known targeted gene knockout method based on the clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9. The researchers codon-optimized the Cas9 protein and designed the guide RNA, then managed to coexpress the gRNA and Cas9 protein in Nannochloropsis oceanica. Next-generation sequencing revealed that the method successfully generated a mutation profile that was dominated by 5-base deletion situated precisely at the gRNA targeted site (Figure 1). Knockout of endogenous nitrate reductase gene via this approach led to growth failure under nitrate (NaNO3), yet uncompromised growth under ammonium (NH4Cl). This is one of the first demonstrations of genome editing in industrial oleaginous microalgae.
The CRISPR/Cas9-based genome-editing method established here expands the reverse genetics toolbox of industrial oleaginous microalgae, and introduces numerous possibilities in the systems and synthetic biology of microalgae-based carbon dioxide capture and conversion. For example, functional probing and validation of every coding- and noncoding-element on the Nannochloropsis genome now become possible. Combined with other platforms in the Single-Cell Center such as single-cell Raman imaging, Raman screening and sequencing, this genome editing platform will enable establishing the genotype and phenotype links with a new level of precision, breadth and throughput. This will lay the foundation for the design and construction of microalgal traits via precision surgery of the genome.
The new study is headed by Professor XU Jian of QIBEBT and supported by funding from MoST (Ministry of Science and Technology of China), National Natural Science Foundation of China (NSFC) and CAS.
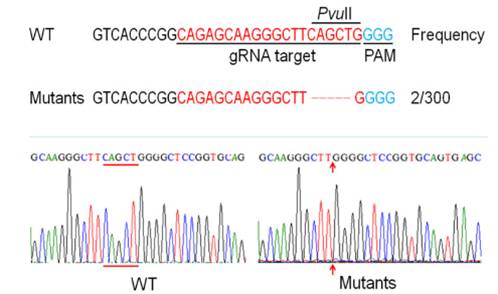
Figure 1. Genotypic validation of the nitrate reductase mutants generated byCRISPR/Cas9-mediated genome-editing(Image by QIBEBT).
Reference:
Genome Editing of Model Oleaginous Microalgae Nannochloropsis spp. by CRISPR/Cas9, Plant Journal, 2016, DOI: 10.1111/tpj.13307; http://onlinelibrary.wiley.com/doi/10.1111/tpj.13307/full
Contact: Prof. XU Jian
Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences
Email: xujian@qibebt.ac.cn