Genetic Engineering of Cellulosome-producing Clostridium Strains for Production of Lignocellulosic Ethanol
Clostridium thermocellum and Clostridium cellulolyticum are typical cellulosome-producing Clostridium strains that can produce ethanol from cellulosic substrates, and have been considered as the most promising candidates for bioconversion of lignocellulose and production of bioethanol via consolidated bioprocessing strategy. However, the wild-type strains contain natural shortcomings including low stress resistance, low yield of ethanol and limited carbon sources, and metabolic engineering has to be performed to meet the requirement of industrialization. Nevertheless, the lack of genetic tools becomes the major bottleneck which delays the application of cellulosome-producing Clostridium strains in utilization of lignocellulose industrially.
In order to achieve the genetic engineering of cellulosome-producing Clostridium efficiently and precisely, Metabolomics Group, led by Prof. CUI Qiu, of Qingdao Institute of Bioenergy and Bioprocess Technology developed a series of genetic tools using mesophilic C. cellulolyticum H10 and thermophilic C. thermocellum DSM1313 as model strains, with which the metabolic engineering of these strains was performed and significantly enhanced the ethanol production. These results have been published on J. Microbiol. Methods, Appl Microbiol Biotechnol and PloS One.
QIBEBT research group firstly developed a custom eletroporator and anaerobic workstation to enhance the stability and efficiency of DNA transformation (Figure 1). We optimized the transformation conditions of both C. cellulolyticum and C. thermocellum and improved the transformation efficiency from 102 to 104/ μg DNA. Subsequently, professor Cui and colleagues expressed an oxygen-independent green fluorescence protein in C. cellulolyticum and C. thermocellum successfully by using different plasmid backbone and thus constructed the fluorescence reporter system to detect the expression of heterologous genes (Figure 2A).
ClosTron system is a direct insertion approach developed on the basis of the mobile group II intron from the ltrB gene of Lactococcus lactis (Ll.ltrB) and has been extensively used in the gene targeting of Clostridium strains. But the application of ClosTron in C. cellulolyticum still contains deficiency. First, C. cellulolyticum harbors a restriction enzyme MspI, which digests heterologous DNA (e.g., transformed plasmids) to protect the cell. Thus, the methylation of the plasmid in advance is required for every transformation, which reduced the efficiency of genetic manipulation dramatically. Second, ClosTron requires the complete plasmid curing of the host cell to exclude the influence of plasmid incapacity in transformation. The frequency of plasmid loss in C. cellulolyticum is very low, and long-term screening process is needed to obtain a plasmid-cured cell chassis, which avoids the successive gene targeting and also decreases the efficiency of genetic manipulation to a great extent. To solve these problems, the researchers disrupted MspI encoding gene in C. cellulolyticum to construct a cell chassis that requires no methylation (Figure 2B) (Cui, Hong et al. 2012). To achieve the successive gene targeting in C. cellulolyticum by use of ClosTron system, a pyrF-based screening system has also been developed (Figure 2C), which can promote the plasmid curing in transformants of C. cellulolyticum and thus improves the ClosTron method (Cui, Zhang et al. 2013).
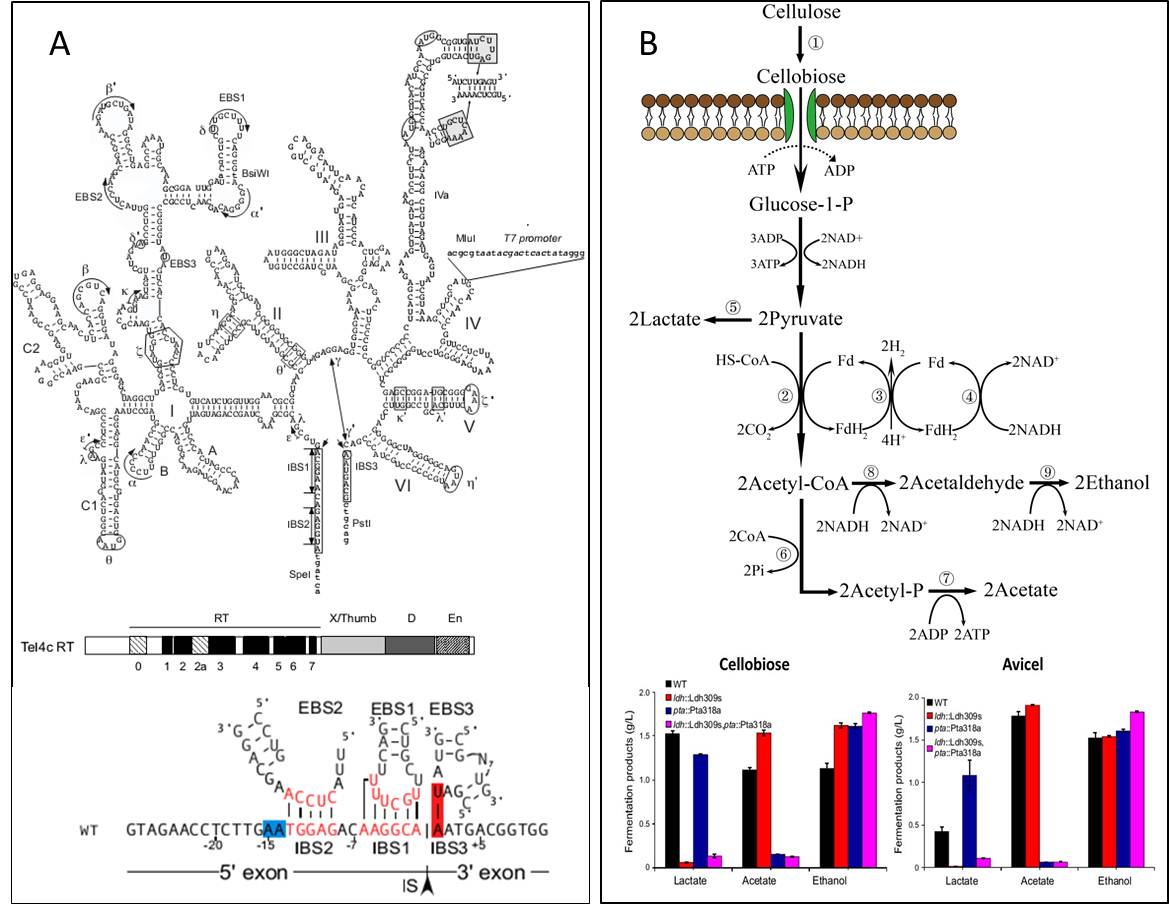
The recent targetrons constructed on the basis of mesophilic group II introns cannot be applied in thermophiles. Hence, prof. CUI cooperated with the research group of Prof. Alan Lambowitz (member of the United States National Academy of Sciences) from University of Texas at Austin and constructed a Thermotargetron system for gene targeting in thermophiles based on a mobile group II intron Tel3c4c from the thermophilic cyanobacterium Thermosynechococcus elongates (Figure 3A), and has been used in C. thermocellum for metabolic engineering (Figure 3B). Besides C. thermocellum, the developed Thermotargetron system can also be used in genetic engineering of other thermophiles (Mohr, Hong et al. 2013).
The genetic tools developed by CUI’s group can be widely applied in the gene disruption and heterologous expression in both mesophilic and thermophilic Clostridium strains. Thus, this study will promote the metabolic engineering of the candidate microorganisms and thus enhance the industrial production of biofuels and related chemicals from lignocellulose.
This work is supported by the National Basic Research Program of China (973-project, grant no. 2011CB707404), the Key Technologies R&D Program from the Ministry of Science and Technology of China (grant no. 2011BAD22B02), and the Instrument Developing Project of the Chinese Academy of Sciences (grant no. YZ201138).

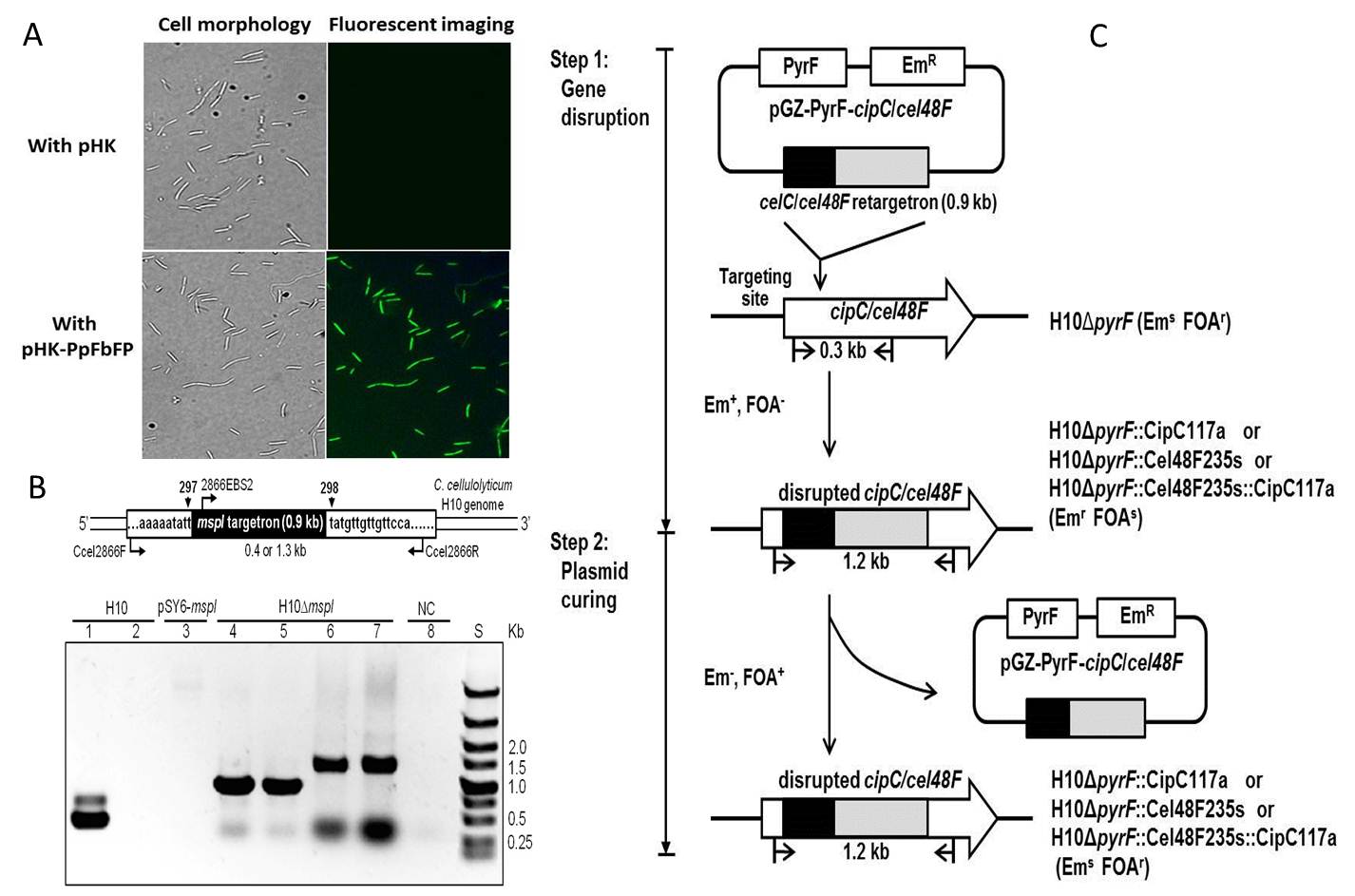
Figure 1. Custom eletroporators and anaerobic workstation for genetic manipulation.
Figure 2. Construction of genetic tools forC. cellulolyticum. A, Confirmation of PpFbFP expression inC. cellulolyticumH10 byin vivofluorescent imaging. B,Construction of the mutant H10DmspIby inactivating the geneccel2866. C, Schematic representation of the 2-step procedure of successive gene disruption using thepyrF-based screening system.
Figure 3. Construction of genetic tools forC. thermocellumand the application in metabolic engineering for ethanol production. A, Tel3c4c group II intron. B, Metabolic engineering for ethanol production inC. thermocellum. (Images by Prof. CUI's group)
Reference:
1. Cui, G. Z., J. Zhang, W. Hong, C. Xu, Y. Feng, Q. Cui and Y. J. Liu (2013). "Improvement of ClosTron for successive gene disruption in Clostridium cellulolyticum using a pyrF-based screening system." Applied Microbiology and Biotechnology.
2. Mohr, G., W. Hong, J. Zhang, G.-z. Cui, Y. Yang, Q. Cui, Y.-j. Liu and A. M. Lambowitz (2013). "A Targetron System for Gene Targeting in Thermophiles and Its Application in Clostridium thermocellum." PLoS ONE 8(7): e69032.
Contact:
Professor CUI Qiu
Email: cuiqiu (AT) qibebt.ac.cn



