Photosynthetic Production of Alka(e)ne Biofuels by Cyanobacteria
Alka(e)ne biofuels are one of the best replacement for fossil-based fuels, because alka(e)nes are drop-in biofuels with the merits such as high-energy density, low hygroscopicity and volatility, and compatibility with current engines and transportation facilities. Considering that cyanobacteria are prokaryotic microbes with photosynthetic capability, it is of great significance to efficiently and directly produce superior and novel biofuels like alka(e)nes in one biological system (cyanobacteria) from solar energy and carbon dioxide.
Researchers from the Metabolic Engineering group at Qingdao Institute of Bioenergy and Bioprocess Technology (QIBEBT), Chinese Academy of Sciences (CAS), have recently made a series of progress in biosynthesis of cyanobacterial alka(e)nes at the multi-scale study with gene, protein and cell.
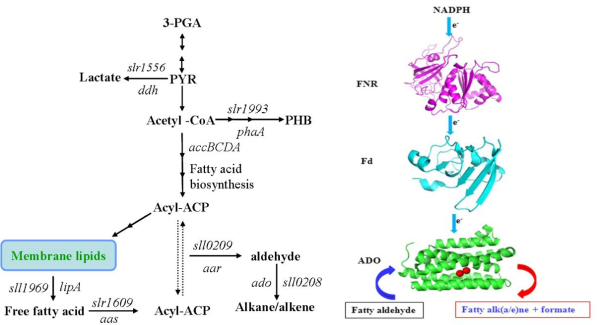
At the gene level---The hydrocarbon compositions of 19 freshwater cyanobacterial species distributed among 13 genera were analyzed. It was found that branched-chain alkanes were limited predominantly in filamentous species but rarely in unicellular species. Phylogenetic analysis using traditional small-subunit ribosomal RNA (16S rDNA) of these strains presented clustering similar to their hydrocarbon production profiles. Acyl–acyl carrier protein reductase (AAR) and aldehyde deformylating oxygenase (ADO) are two key enzymes involved in the biosynthesis of hydrocarbons in cyanobacteria. A comparison of phylogenies revealed that the topology of 16S rDNA showed a general congruence with that of AAR but not with that of ADO. (Applied Energy, 2013, 113, 383-393)
At the protein level---The potential endogenous reducing system including ferredoxin (Fd) and ferredoxin-NADP+ reductase (FNR) to support Aldehyde deformylating oxygenase (ADO) activity was identified in Synechococcus elongatus PCC7942. ADO and the reducing system from Synechococcus elongatus PCC7942 were cloned, overexpressed, and characterized. The cognate electron transfer system supported greater ADO activity than the surrogate (from spinach) and chemical ones. Importantly, kcat value of ADO 1593 using the homologous reducingsystem is 2.7-fold higher than the chemical one. (Biotechnology for Biofuels, 2013, 6:69)
At the cell level---A series of Synechocystis sp. PCC6803 mutant strains have been constructed and confirmed. Alka(e)ne content in a Synechocystis mutant harboring alkane biosynthetic genes over-expressed in both slr0168 and slr1556 gene loci (LX56) was 1.3% of cell dry weight, which was enhanced by 8.3 times compared with wildtype strain (0.14% of cell dry weight) cultivated in shake flasks. Both LX56 mutant and the wildtype strain were cultivated in column photo-bioreactors, and the alka(e)ne production in LX56 mutant was 26 mg/L (1.1% of cell dry weight), which was enhanced by 8 times compared with wildtype strain (0.13% of cell dry weight). (Biotechnology for Biofuels, 2013, 6:69)
For the development of new method for genetic engineering of cyanobacteria---A new screening method free of antibiotics for genetic engineering of cyanobacteria was established using the FLP/FRT recombination system, and antibiotic marker recycling in cyanobacteria was achieved. This has laid the foundation for genetic modification of cyanobacteria through multiple genes and integration sites, and further significantly improving biosynthetic efficiency of cyanobacterial alka(e)nes. (Applied Microbiology and Biotechnology, 2013, doi: 10.1007/s00253-013-4837-6)
The research work was supported by National Basic Research Program of China (973), National Science Foundation of China, Chinese Academy of Sciences, and Boeing Company.
 |
|
Fig1. Biosynthetic pathway of cyanobacterial alka(e)nes and representative electron transfer of ADO-catalyzed reaction |
|
|
|
Fig 2. Analysis of hydrocarbon compositions of representative cyanobacterial strains |
References:
Contact:
Prof. LU Xuefeng
E-mail: lvxf (AT) qibebt.ac.cn