QIBEBT Reveals Cellulose Degradome of Clostridium cellulolyticum
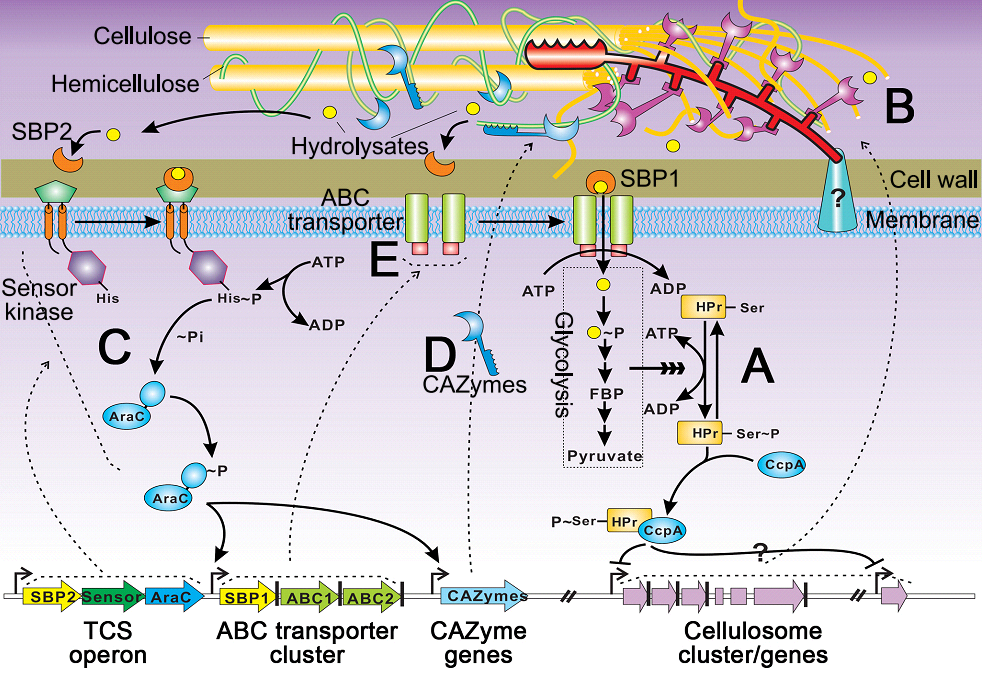 |
| Figure 1. The Cellulose Degradome ofClostridium cellulolyticum. (Image by Prof. XU Jian's Group) |
Reference:
In addition, the team discovered that glucose, which is not a preferred diet of C. cellulolyticum, actually promotes cellulolysis. Glucose achieves the promoting effect by inducing cellulase transcription at low concentrations while by promoting cell growth at high concentrations. Due to their complementarity in preferred substrates and their distinction in functional roles along the reaction chain of cellulose utilization, C. cellulolyticum and the non-cellulolytic and glucose-preferring organisms (e.g., Zymomonas mobile and Saccharomyces cerevisiae) can potentially serve as ideal cellulolytic partners for the Consolidated BioProcessing (CBP) of lignocellulose.
This work was led by Prof. XU Jian, group leader of the Function Genomic Group of CAS-QIBEBT. This work was published online in Biotechnology for Biofuels (2013) 6:73 (doi:10.1186/1754-6834-6-73).
Structure and regulation of the cellulose degradome in Clostridium cellulolyticum, Biotechnology for Biofuels (2013) 6:73 (doi:10.1186/1754-6834-6-73). http://www.biotechnologyforbiofuels.com/content/6/1/73
Lignocellulosic biomass is the most abundant biopolymers on earth, yet its recalcitrance to enzymatic hydrolysis has hampered its exploitation for renewable bioenergy and biomaterials. Cellulolytic bacteria such as Clostridium cellulolyticum, which digest cellulose via a cell surface-attached extracellular enzymatic complex called the cellulosome, represent a major paradigm for efficient biological degradation of cellulosic biomass. However, the genome-wide metabolic and regulatory networks underpinning bacterial cellulose degradation remain poorly understood.
From the Functional Genomics Group of Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences (QIBEBT), Dr. XU Chenggang, PhD student HUANG Ranran and colleagues proposed a molecular model of the “Cellulose Degradome” for one cellulolytic model organism Clostridium cellulolyticum by analyzing its complete genome and comparing its transcriptomes and extracellular proteomes. The model revealed a “core” set of CAZymes required for degrading cellulose-containing substrates, as well as an “accessory” set of CAZymes required for specific non-cellulose substrates. The core and accessory CAZymes are respectively transcriptionally regulated by a Carbon Catabolite Repression (CCR) mechanism and two-component systems (TCSs). The CCR-mediated monitoring of cellular needs for energy and the TCS-mediated sensing of environmental substrate-availability likely ensure both sensitivity to environmental nutrients and the efficiency of cellulose degradome.Thus these findings can serve as a blueprint for constructing potent cellulase systems (in vitro or in vivo) optimized for the targeted substrate.